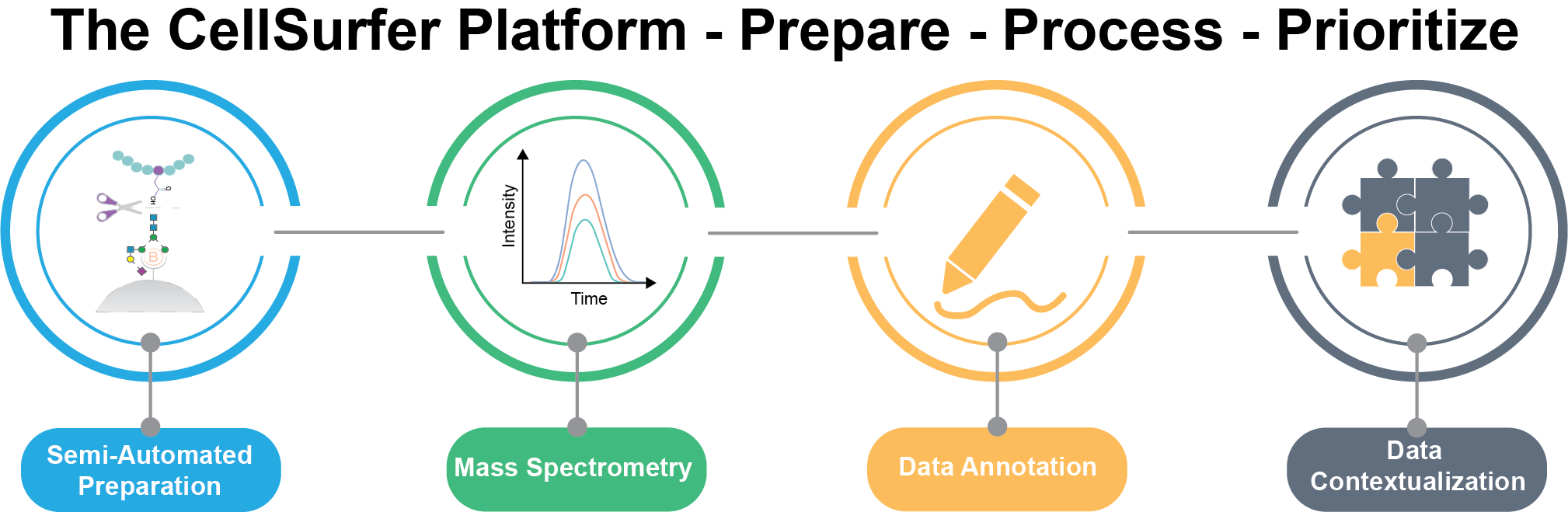
CellSurfer: A platform for discovery of cell surface N-glycoproteins from small sample sizes
CellSurfer integrates efficient sample handling and streamlined data analysis workflows for rapid, quantitative discovery of the cell surface N-glycoproteome.
Prepare:
Microscale cell surface capture (uCSC) enables discovery of cell surface N-glycoproteins from 100 - 1000 ug total cellular peptide (<0.3 - 5 million cells per experiment) and can be completed in 45 hours with minimal human intervention.
Process:
Veneer rapidly curates search engine results and provides enhanced functional annotations to facilitate biological interpretation. Veneer enables consistent interpretation and reporting of μCSC and related datasets
Prioritize:
CIRFESS, SurfaceGenie and Protter are combined to rapidly prioritize candidate markers and assess and visualize surfaceome coverage and extracellular domains for downstream applications
The CellSurfer platform expedites discovery, analysis, annotation, and candidate prioritization for downstream validation of cell-type or condition-specific markers.
CellSurfer is applicable to nearly any mammalian cell type, including sample limited primary cells, for the discovery and quantification of cell surface N-glycoproteins to support the development of immunophenotyping strategies, identification of drug delivery targets, and for studying biological and disease processes.
We are actively applying CellSurfer to develop cell surface markers for stem cell derived cardiomyocytes, mapping the human heart surfaceome in a cell-type specific approach, studying advanced heart failure, and for developing new insights into human development and disease related to blood cancers, viral infection, and pancreatic cells.